convert PDB file to Ansys key points and lines
convert PDB file to Ansys nodes
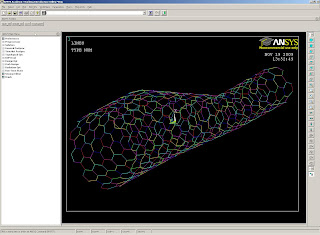
To use these scripts:
1. create a protein data bank file of your structure
2. name it test.pdb and save it in a folder: C:\xyz
3. make sure you have python installed
4. and to make things easier for development and use, grab the free version of Wingware IDE 101
5. download the two scripts and open them in Wing
6. run the scripts
The output file will be in the xyz directory. To load them in Ansys: File > Read input from...

6 comments:
Ha! You use Py too :)
But when processing data,I think awk is easier, so better!
Hi,
Many many thanks for this great work.
I followed the way you suggeted to convert PDB file into ANSYS format. When I import the text files, only nodes are visible. Is there any way get the connectivity (e.g., using beam element etc.)
Seeking your expert advice,
Regards,
Rajib
try the other script
Tom
hi
very tanx for yr script
but i have two problem:
1_i followed the way u suggested but i see only nodes
2_ also i cant see several last nodes or several nodes are omitted
i need yr help & best regard every body can help me.
u can send email: jaavvaaddii@yahoo.com
I can figure out this problem.
1.After all the steps.
2.Close the Python
3.Reopen the text of the atom.
I hope this will be helpful.
hello sir,i am trying to get first PDB file from nano tube modeler software but its giving me message like export functions disabled in unregistered version ,how can i solve this problem can u tell me
thank u
Post a Comment